Analysis of single-nuclei RNA sequencing data
This is a large scientific study in which I analyzed snRNA-seq data from post-mortem human striatal samples to establish a taxonomy of striatal inhibitory neurons.
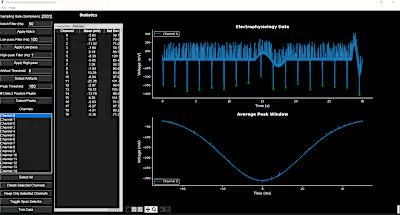
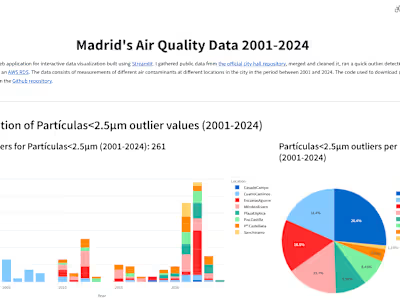
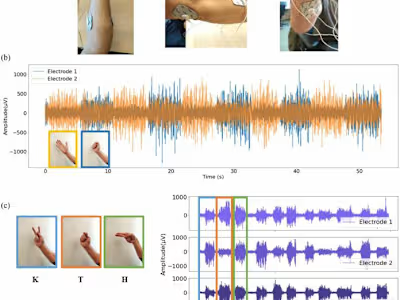
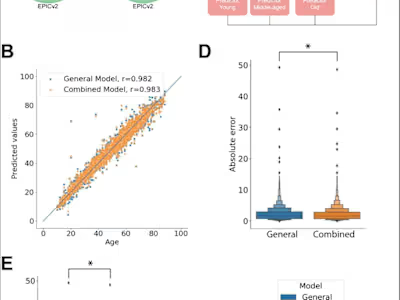
I performed and end-to-end analysis, from fastq to the final publication figures. The analysis included data demultiplexing and alignment of sequencing reads using Illumina's CellRanger and count data processing with Scanpy and several other Python libraries.
To obtain meaningful results from the data on this project, I made extensive use of dimensionality reduction methods, unsupervised learning and statistical testing.
You can read the whole story in the peer-reviewed article linked below.
Like this project
Posted Aug 21, 2024
Analysis of scRNA-seq data from human striatal samples. Used in a scientific project to establish a taxonomy of human striatal inhibitory neurons.
Likes
0
Views
9